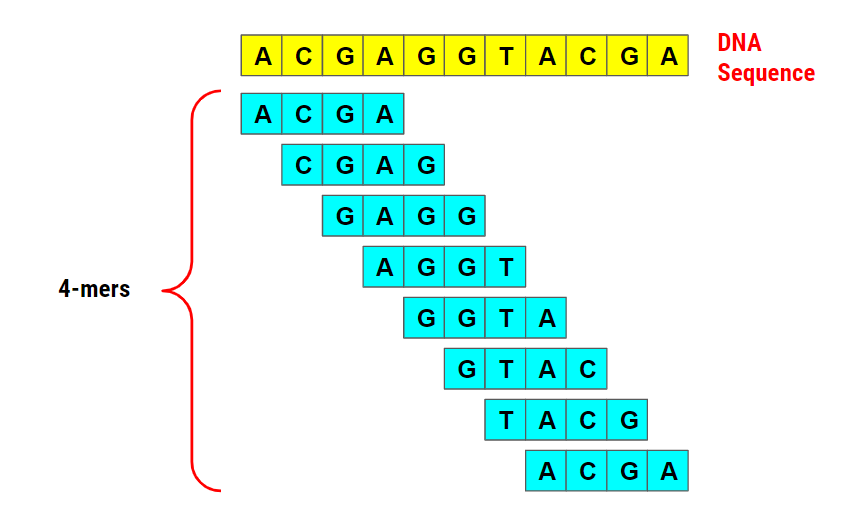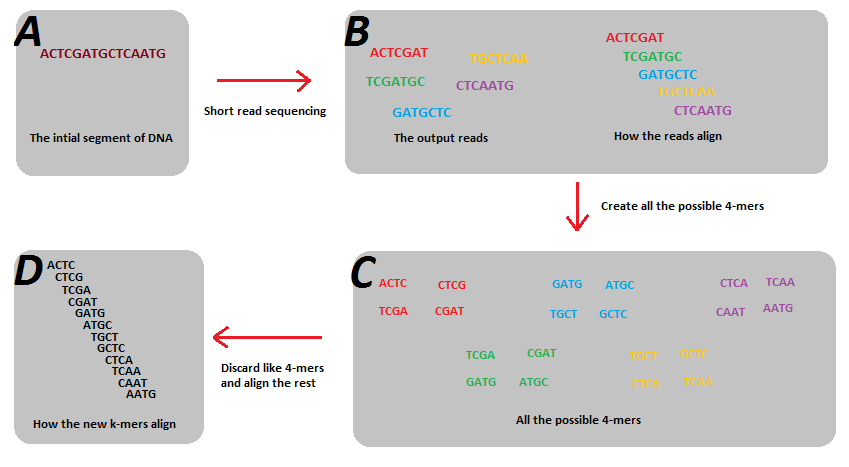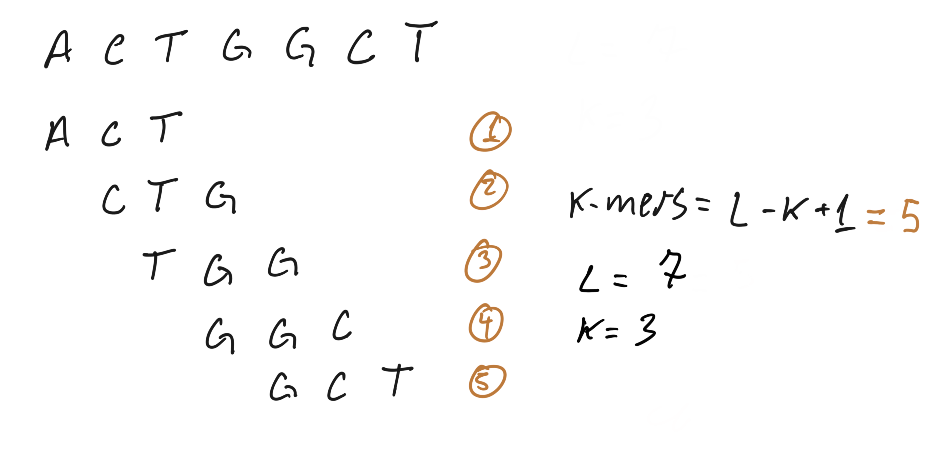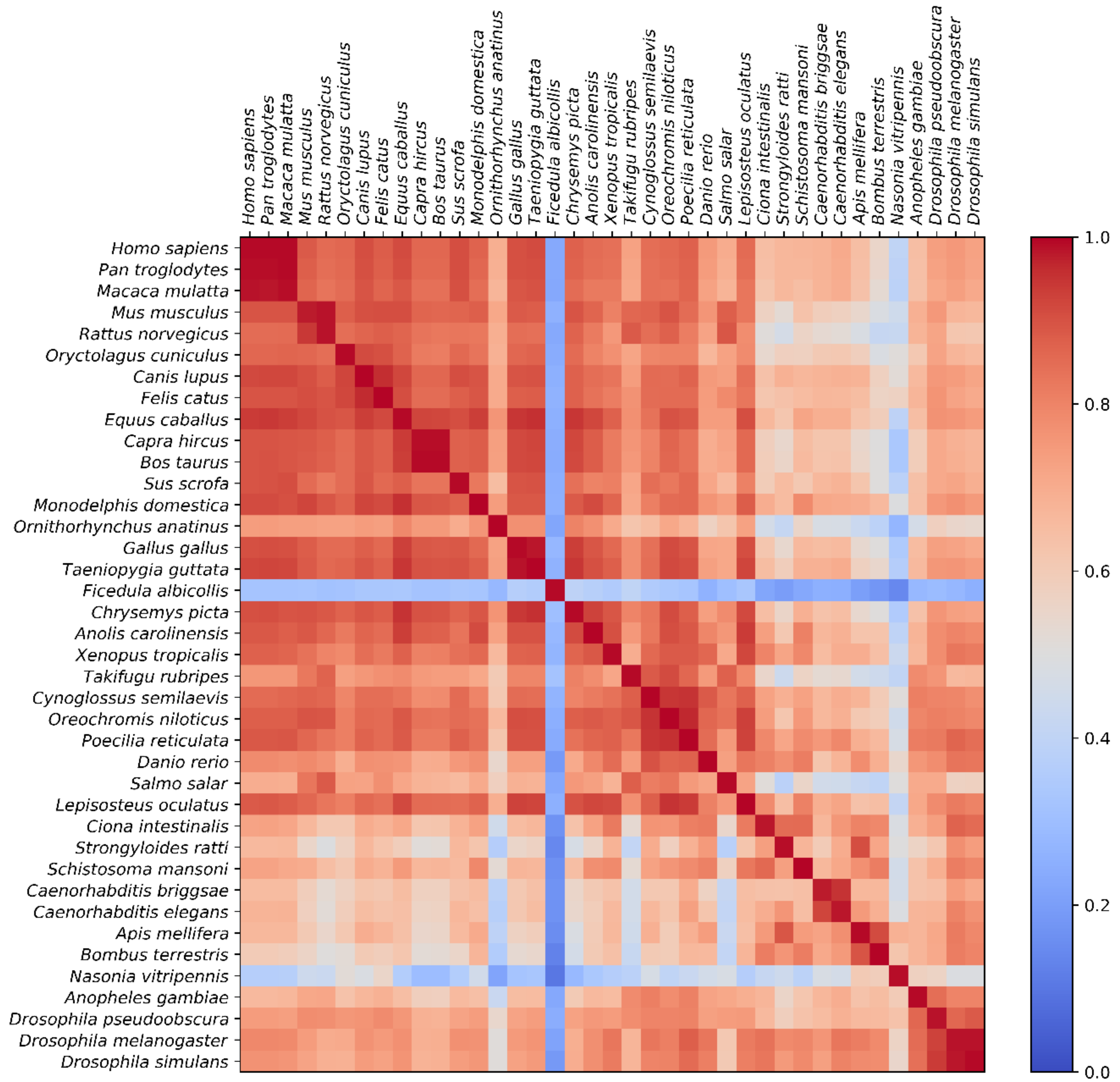
Genes | Free Full-Text | Conservation of k-mer Composition and Correlation Contribution between Introns and Intergenic Regions of Animalia Genomes
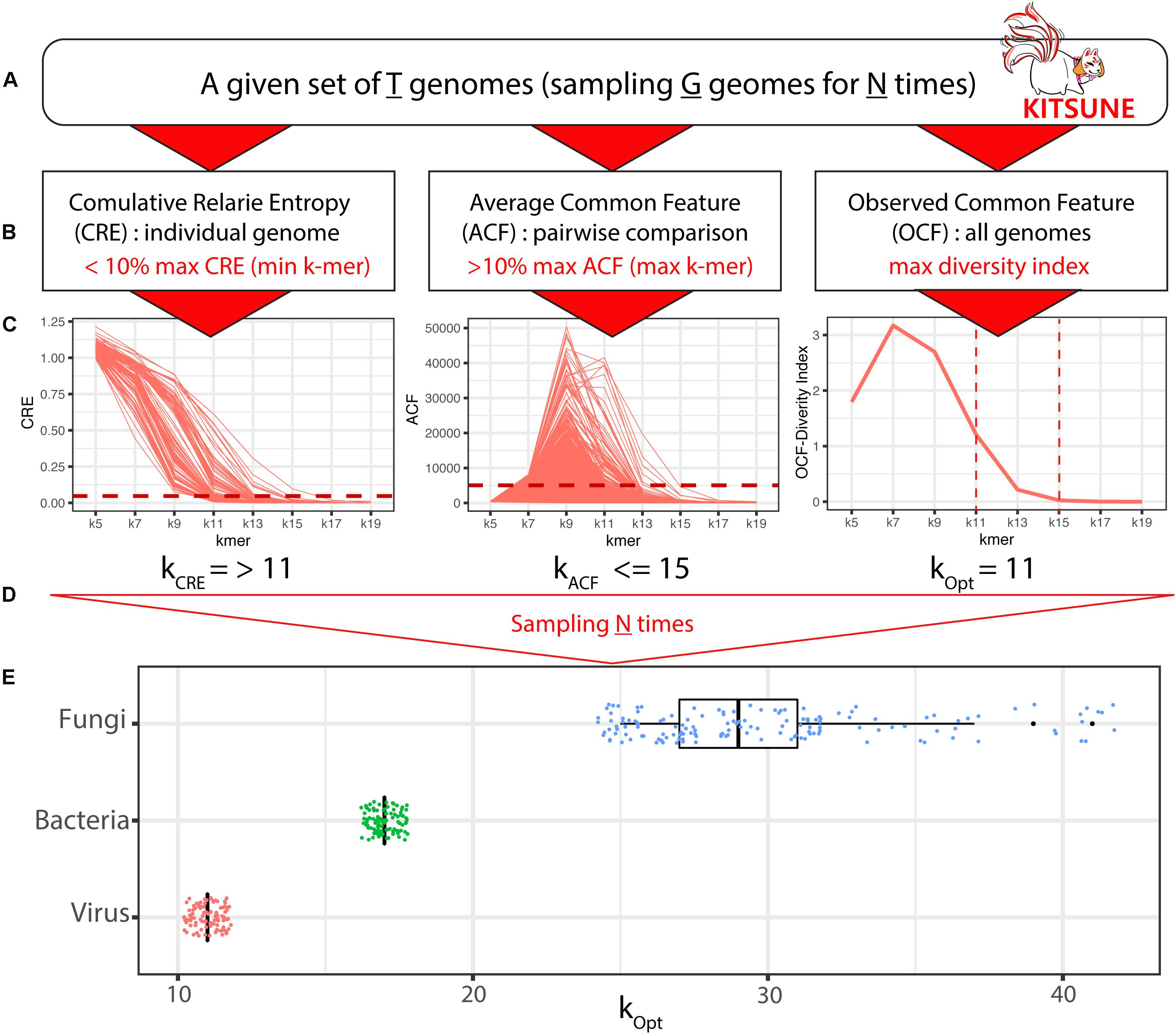
Frontiers | KITSUNE: A Tool for Identifying Empirically Optimal K-mer Length for Alignment-Free Phylogenomic Analysis
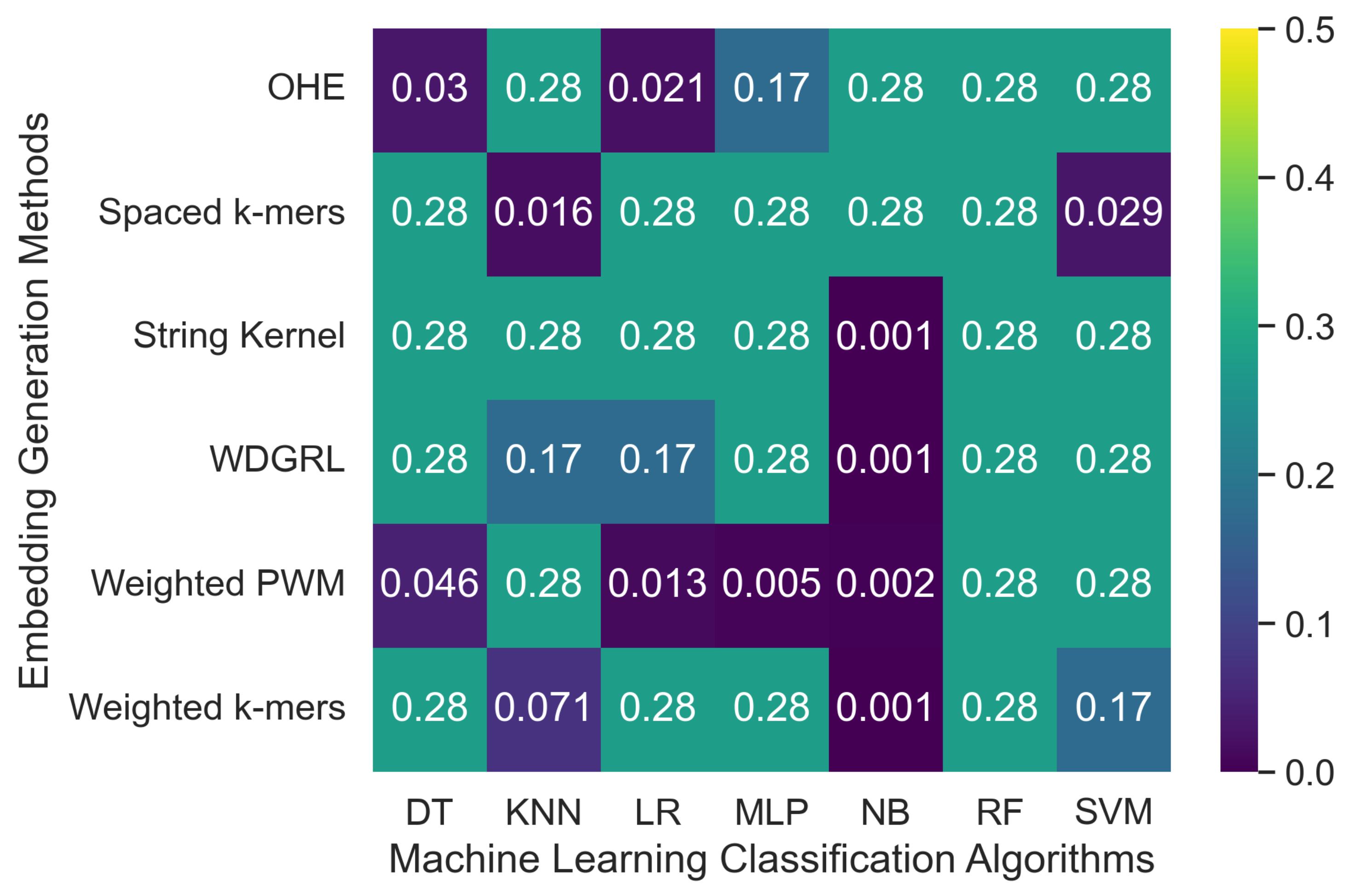
Biomolecules | Free Full-Text | Assessing the Resilience of Machine Learning Classification Algorithms on SARS-CoV-2 Genome Sequences Generated with Long-Read Specific Errors
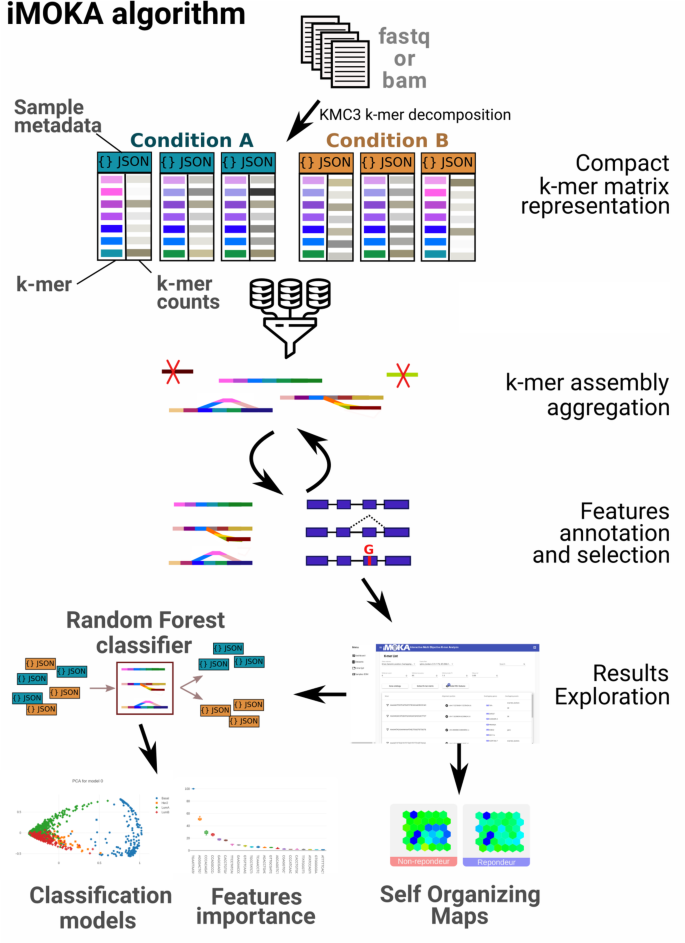
iMOKA: k-mer based software to analyze large collections of sequencing data | Genome Biology | Full Text

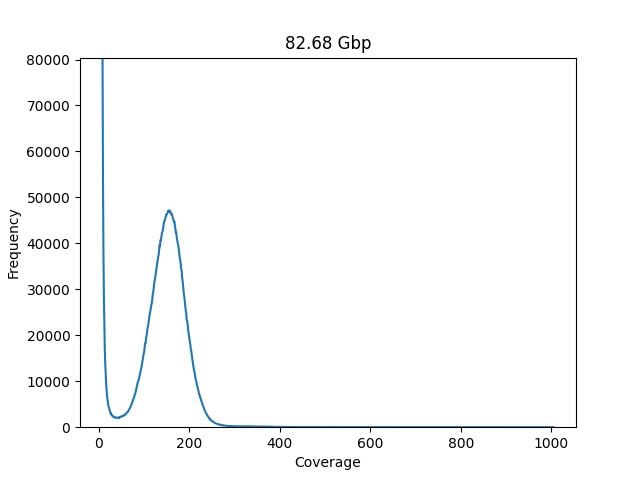
Genome Toolkit. Part 3: building statistical data (k-mer frequency) | by rebelCoder | Python in Plain English
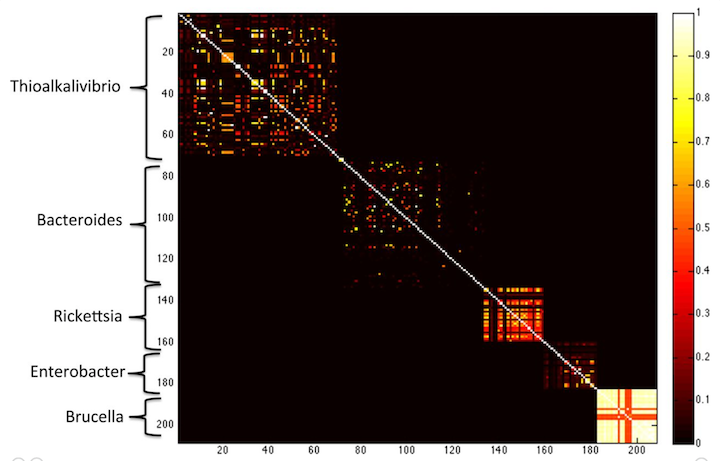
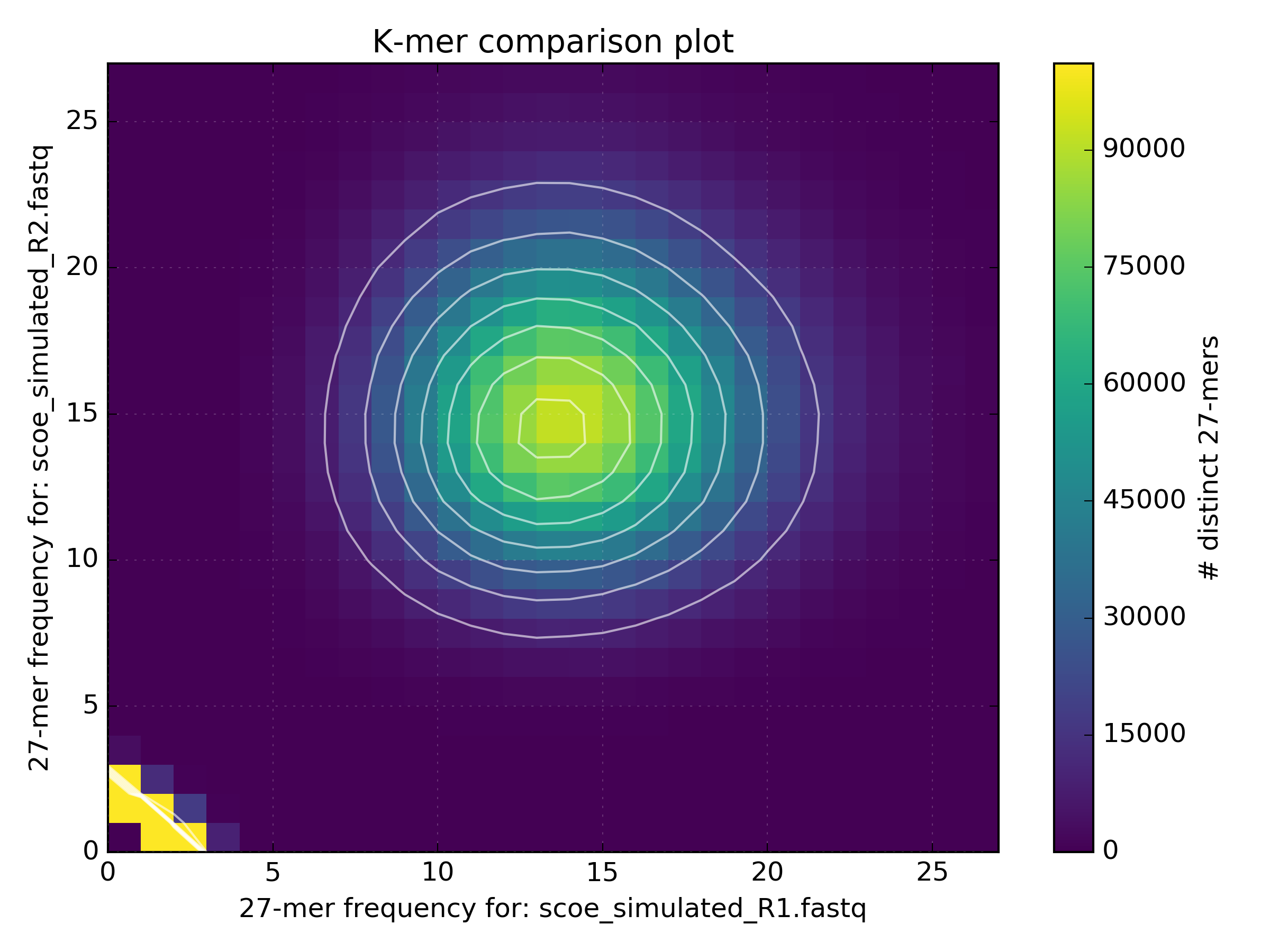
K-mers, k-mer specificity, and comparing samples with k-mer Jaccard distance. — angus 6.0 documentation
GitHub - MindAI/kmer: A Python code for generating k-mer features for a set of DNA/ genomic sequences.